Hello,
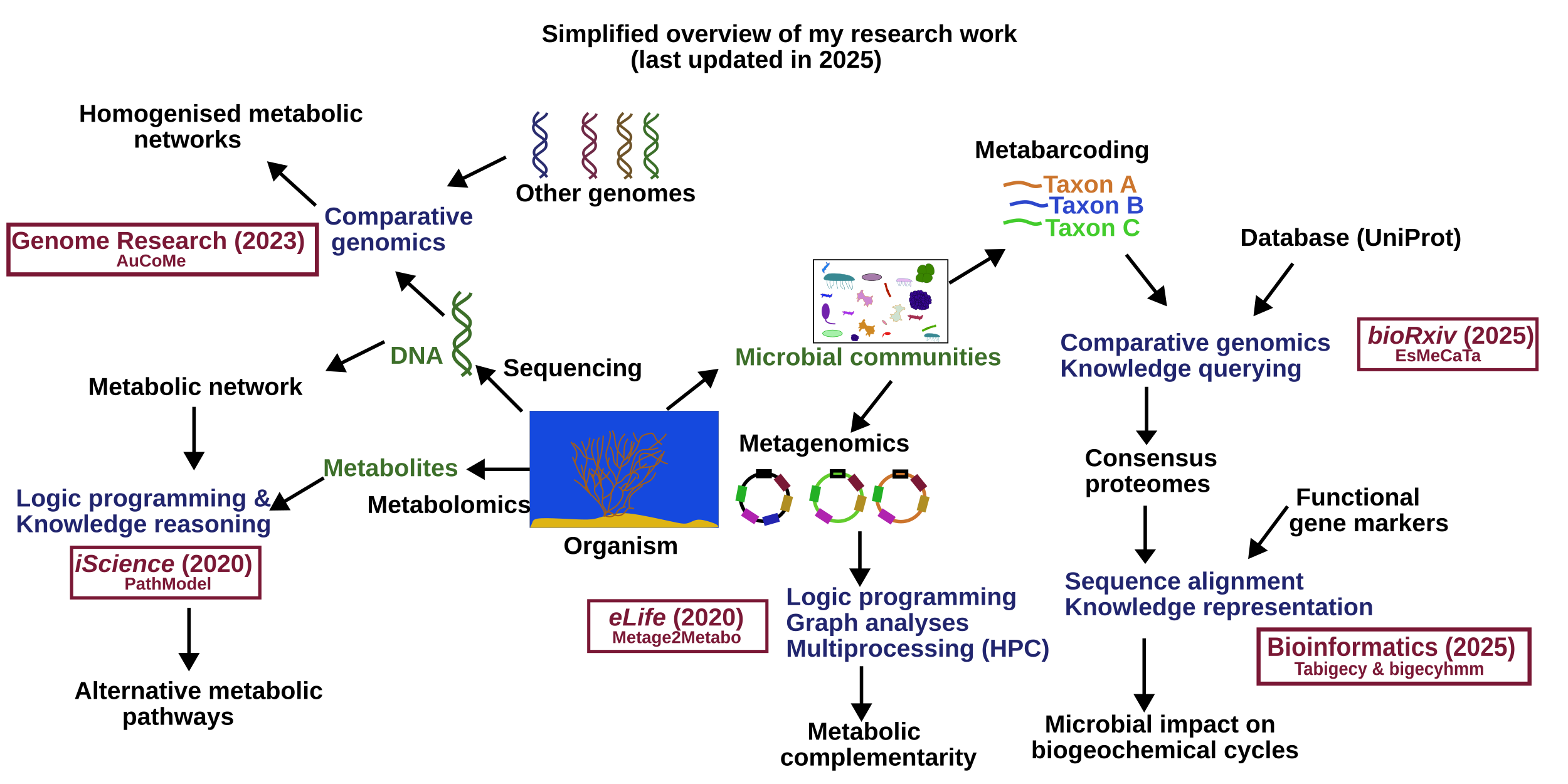
I am a Post-doctoral fellow working on methods to study the metabolism of organism and microbial community in the MICROCOSME Team. My work is at the interface between biology and computer science to better understand different organisms (algae, microbial communities from biogas reactor or underground reservoirs). It is achieved by combining several domains: bioinformatics methods to make predictions from DNA sequences, metabolic predictions with system biology.
Projects
- HyLife-CETP project: Assess microbial risks on hydrogen storage in Europe
Popularisation of my work subject
Metabolism is the set of biochemical reactions that allow the growth of an organism. Biochemical reactions are conversion of substance into other substance through an energy change. In metabolism, most of the reactions are catalysed (meaning increasing their rate of conversion) by specialised proteins called enzymes.
To grow, an organism retrieves nutrients (substances) from their environment, degrades them to generate energy (process called catabolism) and uses this energy to produce other metabolites required for their growth (process called anabolism).
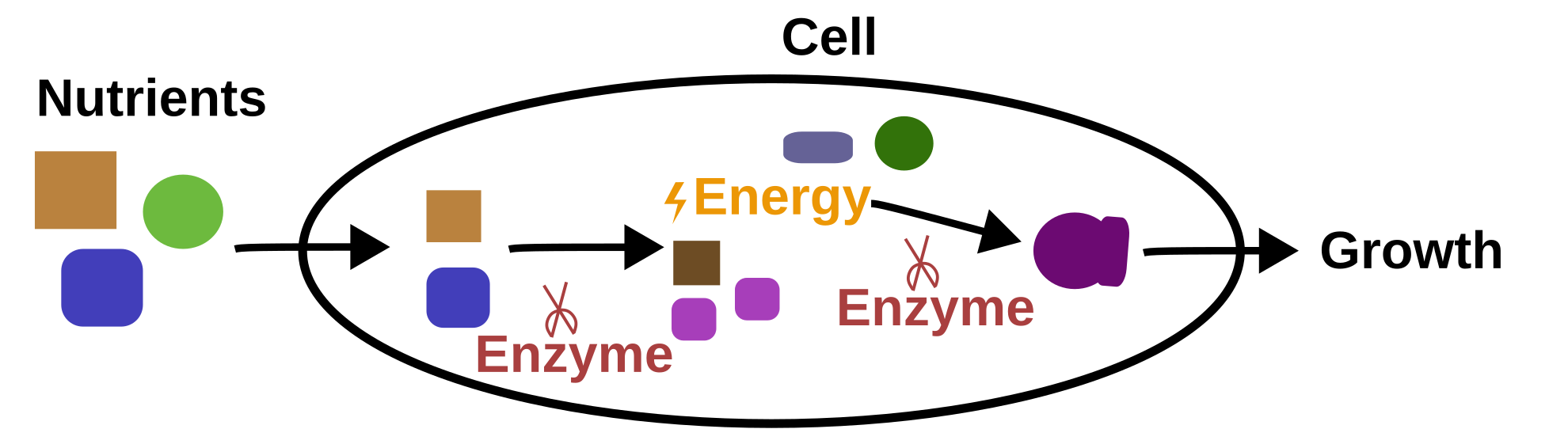
To understand the metabolism of an organism, we need to find which metabolites can be used by it. One possibility is by identifying its enzymes. This is possible because enzymes (like other proteins) are generated by DNA transcription and translation. Inside the DNA sequence, there are regions (called genes) that can be transcripted and translated into proteins (process often simplified as coded).
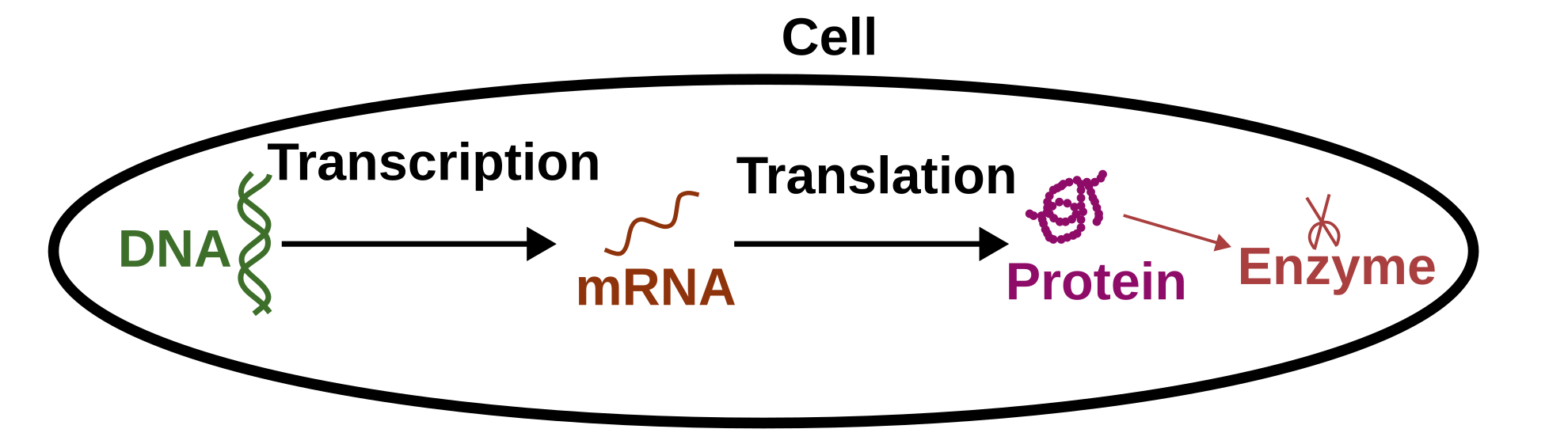
So if we have access to the genome (full DNA sequences of an organism), it is possible to have an idea of its metabolic potential by identifying the genes coding for enzymes. But only an idea, as not all genes are expressed, not all mRNA are translated, some proteins are not produced and some enzymes are working under specific conditions.